Kruskal's Algorithm
This is more of an addendum to Travelling Salesperson. I quickly made a solution to Minimum Spanning Tree problem. This algorithm runs in \(O(\alpha(E)E\log(E))\) time. This is the Inverse Ackerman function \(\alpha\), and it grows incredibly slow, \(\alpha (2^{2^{2^{65533}}} - 3) = 4\). So the analysis of this algorithm would basically be \(O(E\log(E))\), however our set-up is always a connected graph, so we can just denote it as \(E=n(n-1)/2\), which is ~\(n^2\). And for when \(n=|V|\), our algorithm runs in \(O(\alpha(n^2)n^2 2\log(n))\) which is \(O(n^2\log(n))\). For a more detailed analysis, check Wikipedia’s for a simple to follow one here.
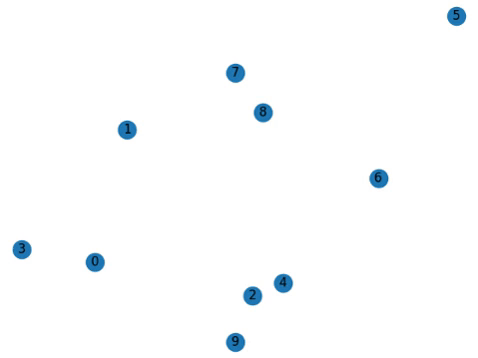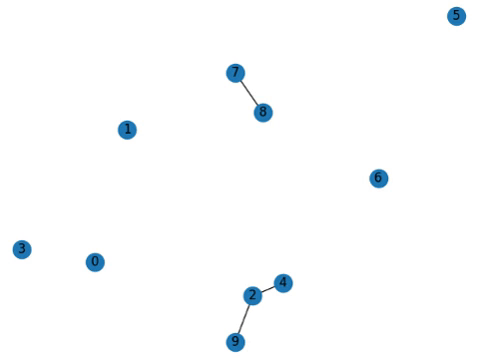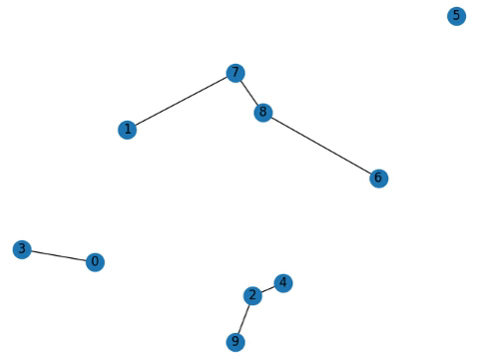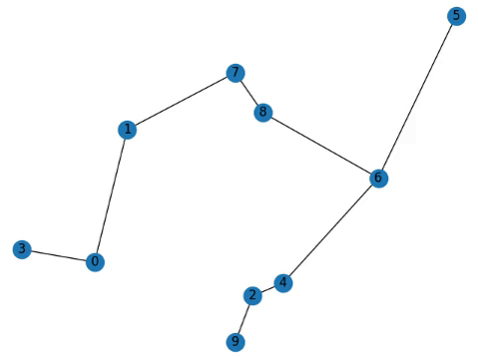
Addendum
I wanted to thank William Edwards, he posed a problem in the USC Codeathon this Spring, and it was the disjoint set data structure. It is brilliantly simple, an incredibly weird run time \(O(\alpha(n))\), and otherwise cool algorithm. It is implemented in a not very forward facing way down below.
Also to show how fast this algorithm is, here’s 100 nodes, which would be practically unsolvable in the Travelling Salesperson Problem.

Source Code
Click here for the source file.
# Copyright 2019 Justin Baum
# Kruskals Algorithm
# kruskals-algo.py
import networkx as nx
import matplotlib.pyplot as plt
import random
#import networkutils as nu
"""
1. Sort all the edges in non-decreasing order of their weight.
2. Pick the smallest edge. Check if it forms a cycle with the spanning tree formed so far. If cycle is not formed, include this edge. Else, discard it.
3. Repeat step#2 until there are (V-1) edges in the spanning tree
"""
def kruskals(matrix):
m = len(matrix)
E = [[i, j, matrix[i][j]] for i in range(m) for j in range(i+1,m)]
E.sort(key = lambda x: x[2])
walk = []
dj = list(range(m))
while len(walk) <= m - 2:
leg = E[0]
E = E[1:]
if find(leg[0],dj) != find(leg[1], dj):
union(leg[0],leg[1],dj)
walk.append(leg)
return walk
### Disjoint Set
def find(i, dj):
while dj[i] != i:
i = dj[i]
return i
def union(i, j, dj):
a = find(i, dj)
b = find(j, dj)
dj[a] = b
def showGraph(V, matrix, walk):
X = nx.Graph()
plt.close()
X.add_nodes_from(V.keys())
E = list(map(lambda x: x[:2], walk))
X.add_edges_from(E)
nx.draw(X, V, with_labels=True)
plt.savefig('fig.png')
## Utils
def gennodes(n, size = 200):
graph = {}
for i in range(n):
x = random.randint(0, size)
y = random.randint(0,size)
graph[i] = (x,y)
return graph
def makematrix(V):
m = len(V)
matrix = [[0 for i in range(m)] for j in range(m)]
for node in V:
for neighbor in V:
point1 = V[node]
point2 = V[neighbor]
matrix[node][neighbor] = int(dist(point1, point2)**0.5)
return matrix
def dist(point1, point2):
return (point1[0] - point2[0])**2 + (point1[1] - point2[1])**2
## Run code
V = gennodes(10)
matrix = makematrix(V)
showGraph(V, matrix, kruskals(matrix))